We give a voice to each single cell of the body to show that diversity is an asset, not a liability.
OPEN POSITION for a computer science honours/PhD student on AI-driven single cell analysis.
Research
Fabilab is driven by people, not academic boundaries. We proudly work on biology, medicine, data science, computer science, artificial intelligence, network science and, sometimes, math. We are enthusiastic leaders in night science and love to generate new ideas and - why not - mix things up. If you like our work, shoot Fabio an email. Some of our current projects are listed below.
Deep evolutionary models of cell identity
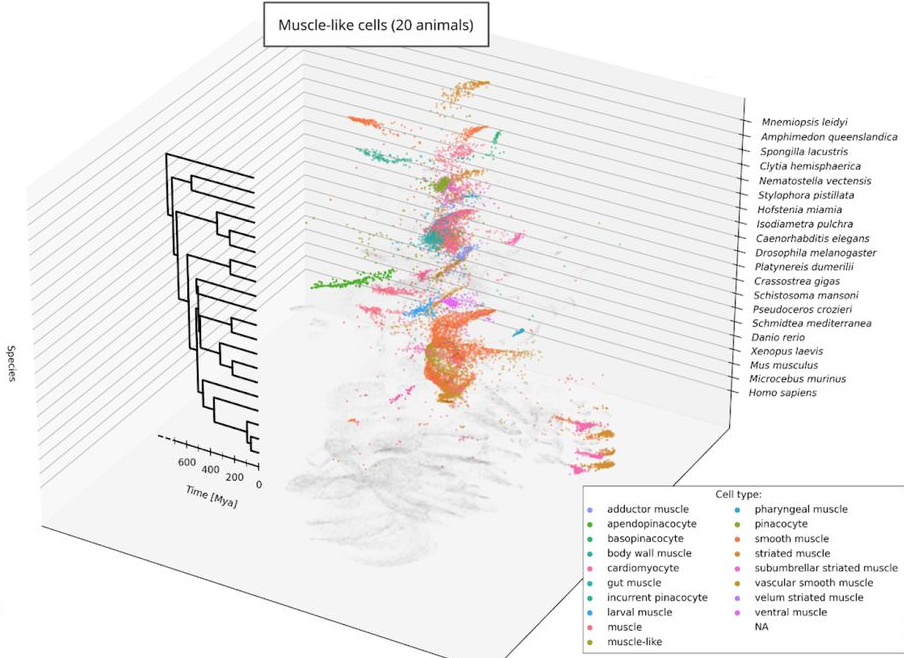
We use artificial intelligence to build universal models of cellular transcriptomes and identity that captures the essence of all eukaryotic cells. We have trained our model to encompass one billion years of cellular evolution, from plants to humans through sponges, worms, and vertebrates. Our model can recapitulate evolution of deeply conserved cell types such as muscle and immune cells, and can predict the identity of cells in new species from which no data is available.
Single-cell measurements across modalities
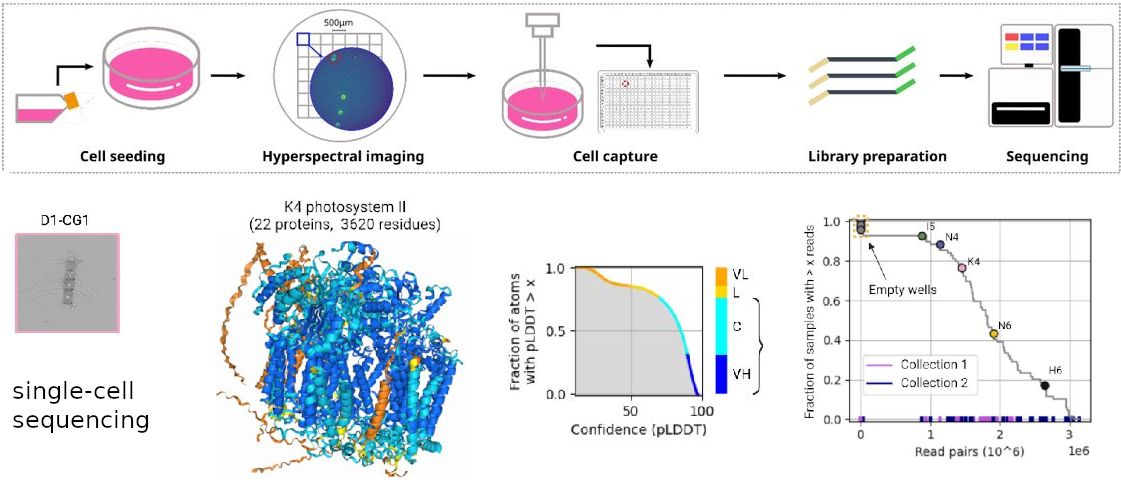
Combining microscopic and transcriptomic measurements on individual cells enables us to examine the relationship between cellular structure and function. We are developing new experimental and computational methods to acquire multimodal single-cell data in both human disease and marine plankton susceptible to climate change.
Virus-inclusive single cell RNA-Seq 2: predictive omics of infection
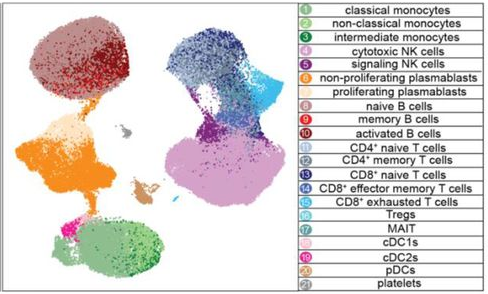
Dengue is the most widespread mosquito-transmitted viral disease with 400 million infections every year. We developnew computational and data science approaches to understand what different parts of the immune system are doing during severe dengue. We identified CD163 as a biomarker for severity in monocytes and patented new antibodies against dengue virus.
Single-cell biomedicine of the neonatal lung
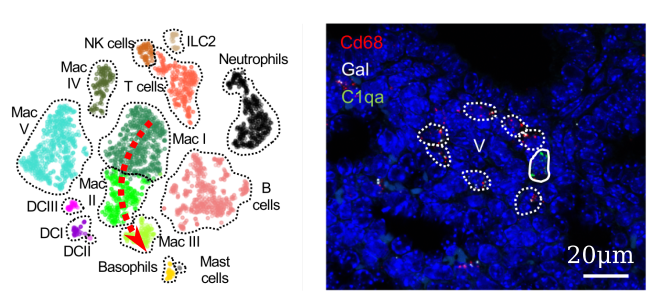
The lung is a very special organ at birth, carrying pathogens and chemically reactive oxygen right into the center of our bodies. Our lab is mapping the staggering cellular diversity characterizing neonatal lungs in terms of gene expression (left) and anatomical location (right). A deeper understanding of this fundamental biological system will help us treat the lung conditions affecting thousands of newborn babies every year, such as bronchopulmonary dysplasia.
Single-cell gene expression with HTSeq 2.0
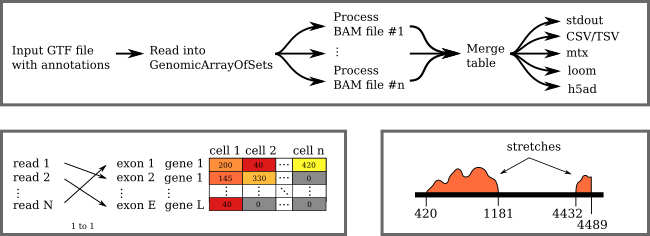
HTSeq is a software library to analyse high-throughput sequencing data in Python. It is particularly popular to quantify gene expression in bulk and single-cell RNA-Seq data via its htseq-count script. We have been maintaining HTSeq for many years and have developed HTSeq 2.0, adding specific support for single cell experiments, exon-level expression, and a dedicated new API to manage islands of dense genomic data sprinkled in data deserts.
Scalable network analysis with igraph
Graphs or networks are an essential mathematical object in modern systems biology. Together with some amazing folks, we develop igraph, a high-performance network analysis tool that underpins many software packages in single cell biology and beyond.
Gene networks regulating leukemic cell state transitions
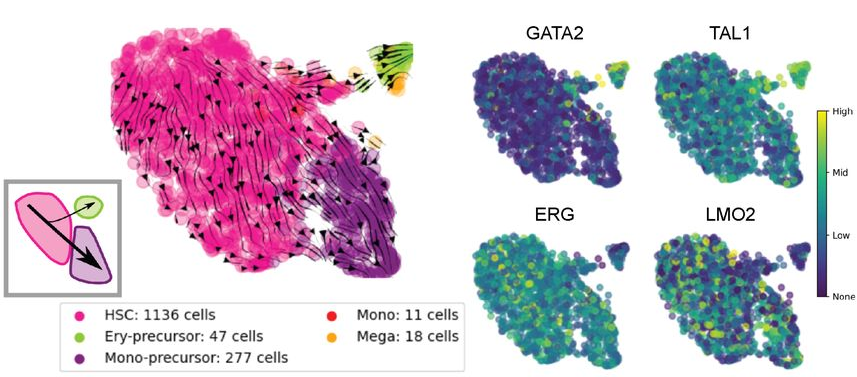
Cancer cells are not equal in the face of therapeutics. Within a single patient, even within a single Petri dish, heterogeneity in gene expression and function distinguishes more stem-like cells, which are more likely to cause relapse, from more differentiated cells. Using our northstar algoritm, we developed data exploration techniques to reveal how the transition between these cancer cell states is regulated and how it can be perturbed chemically, with the vision to reduce relapse in acute myeloid leukemia (AML).